Background
The U-Net architecture was one of the seminal papers in deep learning based medical image segmentation and the architecture became widely used and benchmarked. This model is so successful that its various incarnations dominate this subfield of medical computer vision to this day.
Rarely does a neural network architecture have such a apt name.
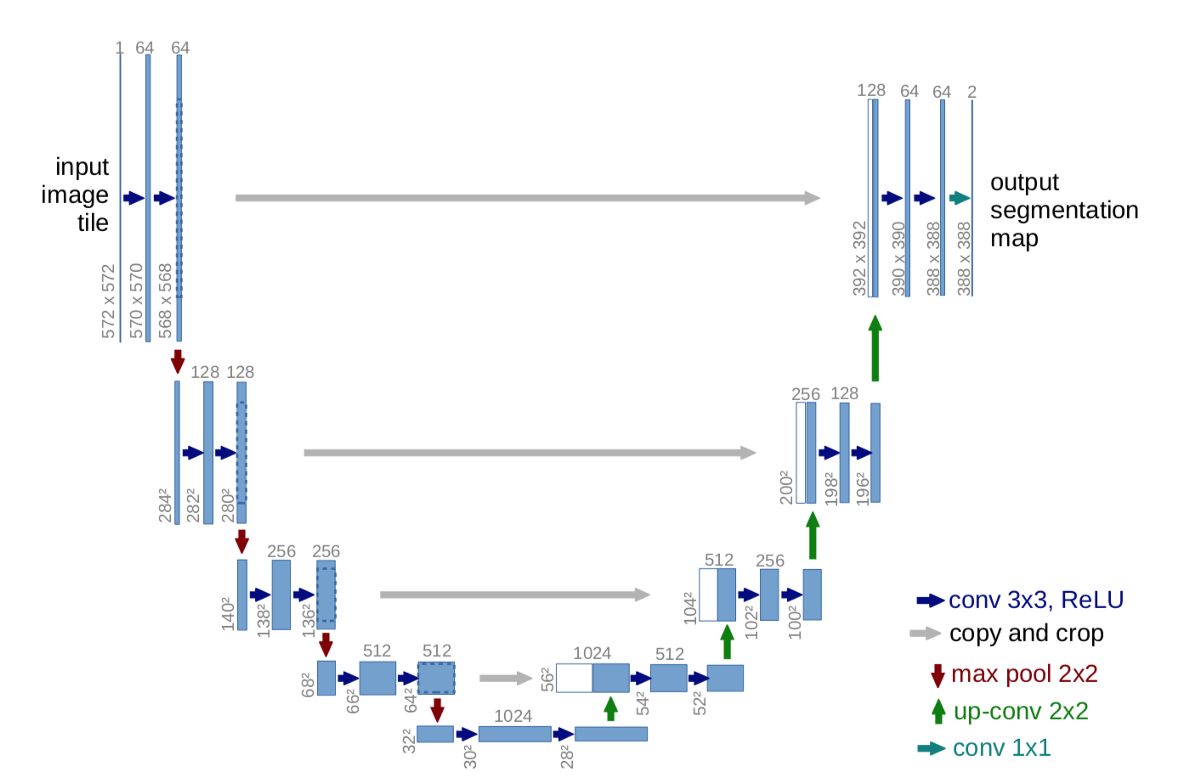
- The task of the network in image segmentation is to identify groups of pixels in a 2D image that “belong together”.
- It achieves this by projecting the input image through a series of convolutional and pooling layers to a low dimensional representation (latent space).
- This is the left part of the letter U (see figure below).
- From this, the network then upsamples the image to the same dimension as the input was and marks each pixel as one of the output classes. Therefore, ideally, the network is patching together nearby pixels into super-cells and large continuous regions, i.e. segmenting the image.
- This is the right part of the letter U.
- The accuracy of this segmentation labelling is checked against the ground truth, which determines the network’s error and therefore the learning process.
The next year, (largely) the same authors released a 3D version of the architecture. As you can imagine, if you have thin 2D slices of a 3D biological (or any physical) structure stacked on top of each other, it doesn’t make much sense to run U-Net on each slice separately. Why? Because you throw out a ton of useful information by not letting the model know that these 2D images are in fact connected in a 3rd dimension and therefore the spatial distribution of class labels is not 2 but 3 dimensional.
This project
For a job interview with a DeepMind, I had to implement the 3D U-Net in TensorFlow from the paper, and train it on the NCI-ISBI 2013 Challenge: Automated Segmentation of Prostate Structures dataset, which consists of 80 patients’ 3D MRI scans from their prostate region.
During this assignment I had to:
- explore the dataset
- get the model to train
- test it on the holdout set
- benchmark it against the paper’s results
- write a report of my work.